Yosef Buganim 1, Dina A Faddah, Rudolf Jaenisch
Affiliations expand
- PMID: 23681063
- PMCID: PMC4060150
- DOI: 10.1038/nrg3473
Free PMC article
Abstract
Conversion of somatic cells to pluripotency by defined factors is a long and complex process that yields embryonic-stem-cell-like cells that vary in their developmental potential. To improve the quality of resulting induced pluripotent stem cells (iPSCs), which is important for potential therapeutic applications, and to address fundamental questions about control of cell identity, molecular mechanisms of the reprogramming process must be understood. Here we discuss recent discoveries regarding the role of reprogramming factors in remodelling the genome, including new insights into the function of MYC, and describe the different phases, markers and emerging models of reprogramming.
Figures
See this image and copyright information in PMC
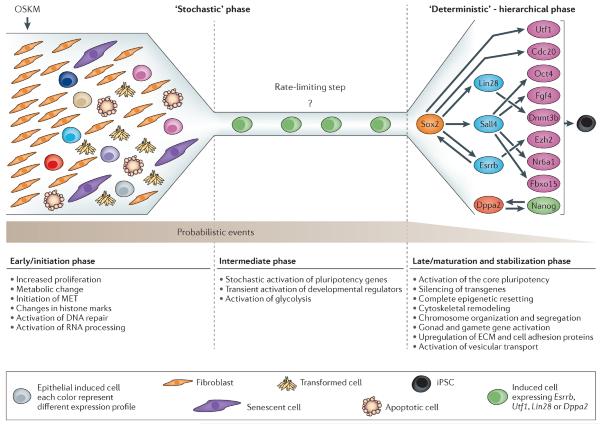
Figure 1. Phases of the reprogramming process
In the model we discuss in this review, the reprogramming process can broadly be divided into two phases: firstly, a long `stochastic’ phase of gene activation; and secondly, a shorter hierarchical more `deterministic’ phase of gene activation that begins with the activation of the Sox2 locus. After a fibroblast is induced with OSKM, it will initiate stochastic gene expression and assume one of several possible fates (such as, apoptosis, senescence, transformation, transdifferentiation or reprogramming). In the early phase, reprogrammable cells will increase proliferation, undergo changes in histone modifications at somatic genes, initiate mesenchymal to epithelial transition, and activate DNA repair and RNA processing. Then the reprogrammable cells will enter an intermediate phase with an unknown rate-limiting step that delays the conversion to iPSCs and contributes to the long latency of the process. In this phase, cells undergo a stochastic activation of pluripotency markers, a transient activation of developmental regulators, and activation of glycolysis. In general the transcriptional changes in this phase are small. In some rare cases, the stochastic gene expression will lead to the activation of “predictive markers” such as Utf1, Esrrb, Dppa2, and Lin28, which then will instigate the second phase that starts with the activation of Sox2. Activation of Sox2 by the “predictive markers” can be direct or indirect and will trigger a series of deterministic events that will lead to an iPSC. In this late phase, the cells eventually stabilize into the pluripotent state in which the transgenes are silenced, the cytoskeleton is remodeled to an ESC-like state, the epigenome is reset and the core pluripotency circuitry is activated–,. In this model, probabilistic events decrease and hierarchical events increase as the cell progresses from a fibroblast to an iPSC.

See this image and copyright information in PMC
Figure 2. OSKM as pioneer factors for remodeling the epigenome
During reprogramming, exogenous OSKM bind enhancers and promoters of fibroblast and ESC genes along with regions that are not occupied by OSKM in ESCs and are not specific to fibroblasts (here called `somatic’). The factors mark the loci that eventually will be epigenetically modified. In general, OSKM bind four different classes of genes. The first class (Fib) contains genes that are important for the identity of the fibroblasts such as Thy1, Postn, Col5a2 and EMT genes like SnaiI, SnaiII and Twist1. The second class (Somatic) contains genes that are bound by OSKM in somatic cells but not in ESCs and are not specific to fibroblasts. This includes apoptotic genes such as p53, genes that are important for proliferative cells, such as cell cycle genes (for example Bub1, Cdc20 and Cdc25c), and metabolic genes such as Pfkl and Gp. The third class (ES-I) contains ESC genes that are activated early in the process such as Fbxo15, Fgf4 and Sall4. The fourth class (ES-II) contains genes that are activated late in the reprogramming process such as Sox2, Nanog and Dppa4. During the early phase of reprogramming, OSKM occupy the enhancers of all classes except enhancers of ES-II genes that contain the heterochromatin mark H3K9me3 and are refractory to the four factors. c-Myc and Klf4 bind promoters of Fib genes and repress their activity while increasing the activation of genes from the Somatic class (shown by the weight of the arrow). As a result, enhancers and promoters from Fib start to lose H3K4me2 while genes from the Somatic class maintain high levels of H3K4me2. OSK act as pioneer factors and occupy the distal enhancer of ES-I genes, which gain de novo H3K4me2 and will initiate expression a few days later. The late phase is less well understood, but it can be speculated that Fib genes become heterochromatic and are silenced while the genes from the Somatic class are highly activated. ES-I genes are highly activated and contain high levels of H3K4me2 and ES-II genes start to lose the H3K9me3 mark, gain H3K4me2 marks and initiate expression. It is reasonable to assume that more pluripotency late factors that are switched on late in reprogramming are needed to open those “blocked” regions. After the silencing of the exogenous factors, all groups are highly expressed except Fib, which remains silenced. The sizes of the ovals representing OSKM indicate their binding preference. For example, c-Myc is a global amplifier of gene expression increasing the transcription at all active promoters, therefore the oval “M” is larger on promoters.
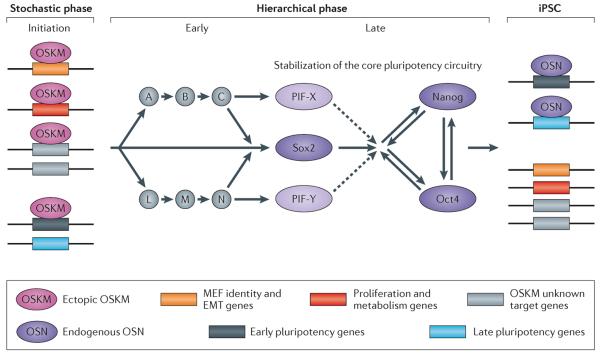
See this image and copyright information in PMC
Figure 3. Model of molecular events that precede iPS formation
In the early phase, ectopic OSKM act as pioneer factors and occupy many genomic regions and help to generate a hyperdynamic chromatin state. OSKM will bind many regions throughout the genome of the fibroblast that are not OSKM targets in ESCs. Among these regions are: genes that determine the identity of the fibroblast, like extracellular components and EMT genes (orange box); genes that promote proliferation and increase metabolism (red box); unknown target genes that facilitate genomic fluidity, i.e., a state that allows rapid changes in transcription (gray box). In addition, OSKM will occupy distal regions of early pluripotency genes (black box); this binding will aid in activating those loci at later stages. A group of late pluripotency genes (blue box) is refractory to OSKM binding in this early phase. In the early hierarchical phase (which is more speculative), early pluripotency genes become activated in rare individual cells and either directly or in a hierarchical manner will instigate a more deterministic process that eventually leads to the activation of Sox2. Sox2 represents one gene of a group of late pluripotency initiating factors (PIFs) that are essential for the activation of the core pluripotency circuitry. Once activated, the endogenous pluripotency proteins Oct4, Sox2 and Nanog (OSN) occupy their target genes and maintain the iPSC state in the absence of the exogenous factors.