Yosef Buganim 1, Dina A Faddah, Albert W Cheng, Elena Itskovich, Styliani Markoulaki, Kibibi Ganz, Sandy L Klemm, Alexander van Oudenaarden, Rudolf Jaenisch
Abstract
During cellular reprogramming, only a small fraction of cells become induced pluripotent stem cells (iPSCs). Previous analyses of gene expression during reprogramming were based on populations of cells, impeding single-cell level identification of reprogramming events. We utilized two gene expression technologies to profile 48 genes in single cells at various stages during the reprogramming process. Analysis of early stages revealed considerable variation in gene expression between cells in contrast to late stages. Expression of Esrrb, Utf1, Lin28, and Dppa2 is a better predictor for cells to progress into iPSCs than expression of the previously suggested reprogramming markers Fbxo15, Fgf4, and Oct4. Stochastic gene expression early in reprogramming is followed by a late hierarchical phase with Sox2 being the upstream factor in a gene expression hierarchy. Finally, downstream factors derived from the late phase, which do not include Oct4, Sox2, Klf4, c-Myc, and Nanog, can activate the pluripotency circuitry.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
See this image and copyright information in PMC
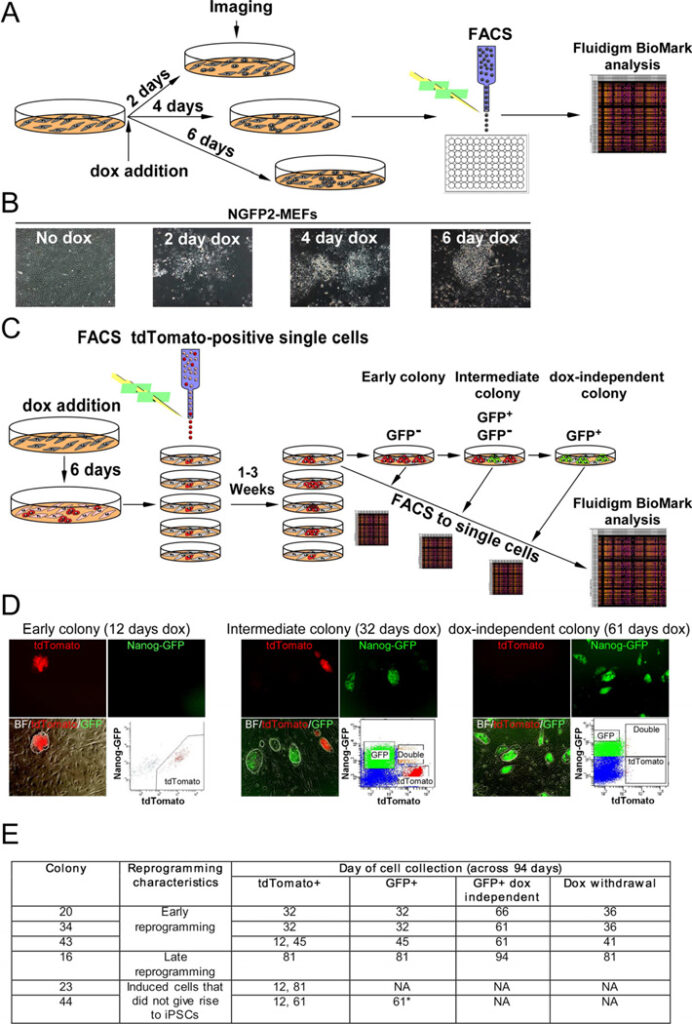
Figure 1. Experimental scheme used to monitor transcriptional profiles of single cells at defined time points during the reprogramming process
(A) Scheme used for single-cell gene expression analysis with Fluidigm. (B) Representative images of NGFP2 MEFS without dox and at days 2, 4, and 6 on dox. (C) Scheme of NGFP2/tdTomato secondary system used to measure single-cell gene expression of clonal dox-dependent (GFP−, GFP+) and independent (GFP+) cells. (D) Representative images and FACS analysis of dox-dependent and independent cells at days 12, 32, and 61 on dox. (E) Six colonies were profiled over the course of 94 days. Colony 44 (starred) contained a few cells with a low level of GFP that were sorted at day 61 and disappeared upon continual passaging and dox-withdrawal. See also Figure S1 and S2.
See this image and copyright information in PMC
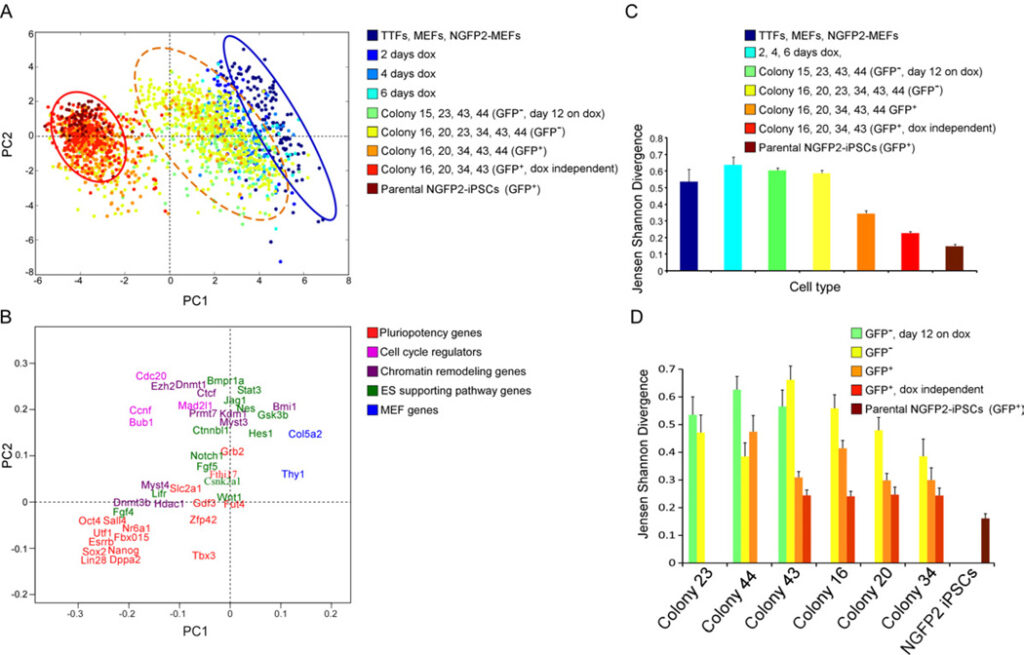
Figure 2. Three reprogramming states
(A) Principal component (PC) projections of individual cells, colored by their sample identification. The blue circle surrounds one population and the red circle surrounds another population. The orange dotted circle surrounds a third intermediate population. (B) PC projections of the 48 genes, showing the contribution of each gene to the first two PCs. The first PC can be interpreted as discriminating between cluster 1 and cluster 2; the second between pluripotency genes and cell cycle regulators. (C-D) Jensen Shannon Divergence analysis of within-group (C) and within-colony (D) variability, colored by the same sample identification as in (A). Error bars represent the 95% confidence interval. See also Figure S3.
See this image and copyright information in PMC
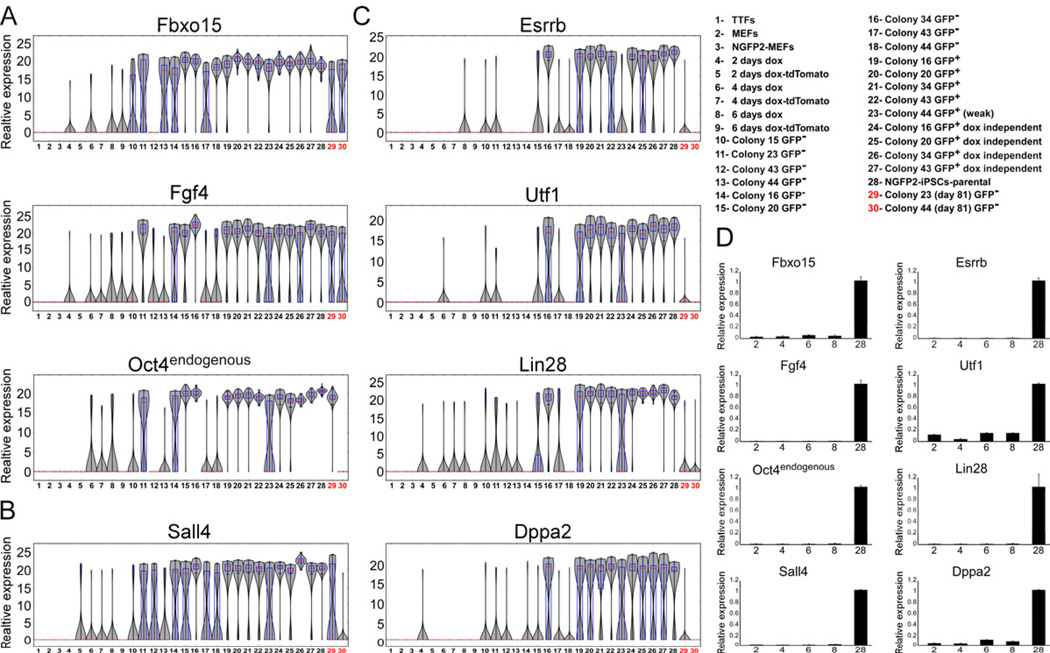
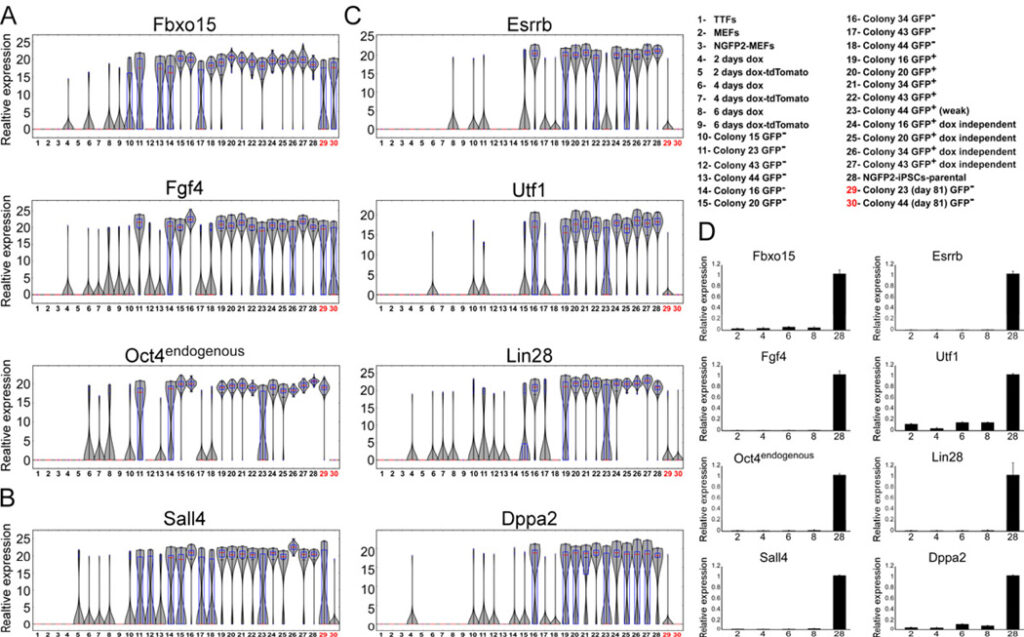
Figure 3. Established early markers are not sufficient to mark cells that will become iPSCs
mRNA expression levels of (A) Fbxo15, Fgf4 and Oct4 (B) Sall4 and (C) Esrrb, Utf1, Lin28, Dppa2 in populations noted in Figure 1 and legend (upper right) are shown in violin plots. Median values are indicated by red line, lower and upper quartiles by blue rectangle, and sample minima/maxima by black line. The two partially reprogrammed colonies (colonies 23 and 44) are marked in red. (D) Quantitative RTPCR of Fbxo15, Fgf4, Oct4, Sall4, Esrrb, Utf1, Lin28, and Dppa2 expression in nonclonal cell populations noted in legend (upper right numbers correspond to x-axis), normalized to the Hprt house keeping control gene. Error bars are presented as a mean ± standard deviation of two duplicate runs from a typical experiment. See also Figure S4 and S5.
See this image and copyright information in PMC
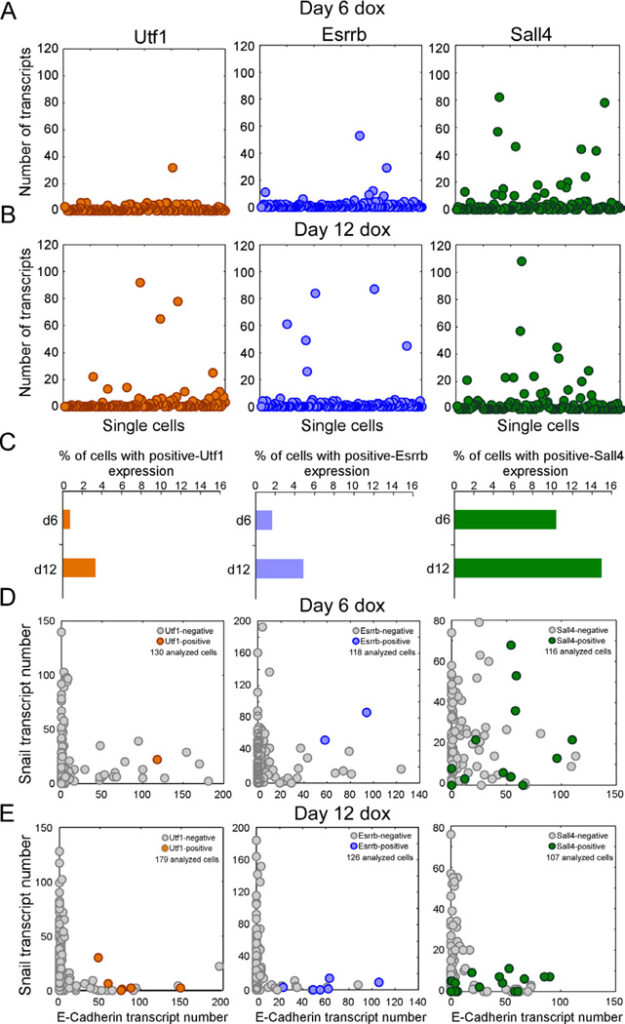
Figure 4. Early markers for reprogramming
(A and B) sm-mRNA-FISH of Utf1 (orange), Esrrb (blue), Sall4 (green) expression in NGFP2 cells at day (A) 6 and (B) 12 on dox. Each cell is represented as a single dot. 120 cells were analyzed for each one of the six plots. (C) Percent of total cell population with high Utf1, Esrrb, and Sall4 at day 6 and day 12. (D and E) sm-mRNA-FISH of Snail vs. E-cadherin expression in single NGFP2 cells at day (D) 6 and (E) 12 on dox. High Utf1 (orange), Esrrb (blue), and Sall4 (green) cells are highlighted. The number of cells analyzed is noted on each plot.
See this image and copyright information in PMC
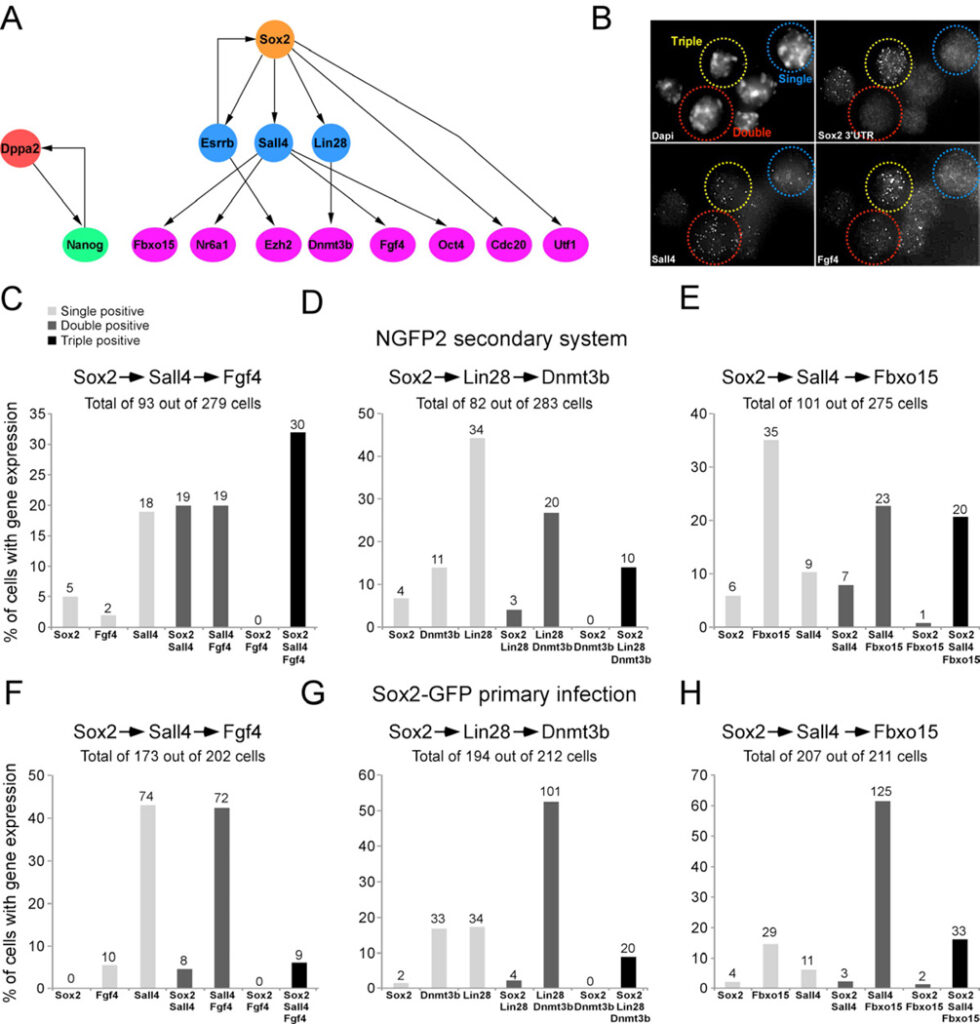
Figure 5. Model to predict the order of transcriptional events in single cells
(A) Bayesian network to describe the hierarchy of transcriptional events among a subset of pluripotent genes. (B) sm-mRNA-FISH representative image of combination in Figure 5C showing a single positive cell (blue, Sall4), double positive cell (red, Sall4/Fgf4), and triple positive cell (yellow, Sox2/Sall4/Fgf4). (C–E) Bar plot of the percent of cells with transcripts, quantified by single molecule mRNA FISH, of single positive (light grey), double positive (dark grey), and triple positive (black) expression in single NGFP2 cells at day 12 on dox and in (F-H) single primary infected Sox2-GFP cells at day 12 on dox. The numbers of cells in each category is indicated on top of each bar. See also Figure S6 and Table S5.
See this image and copyright information in PMC
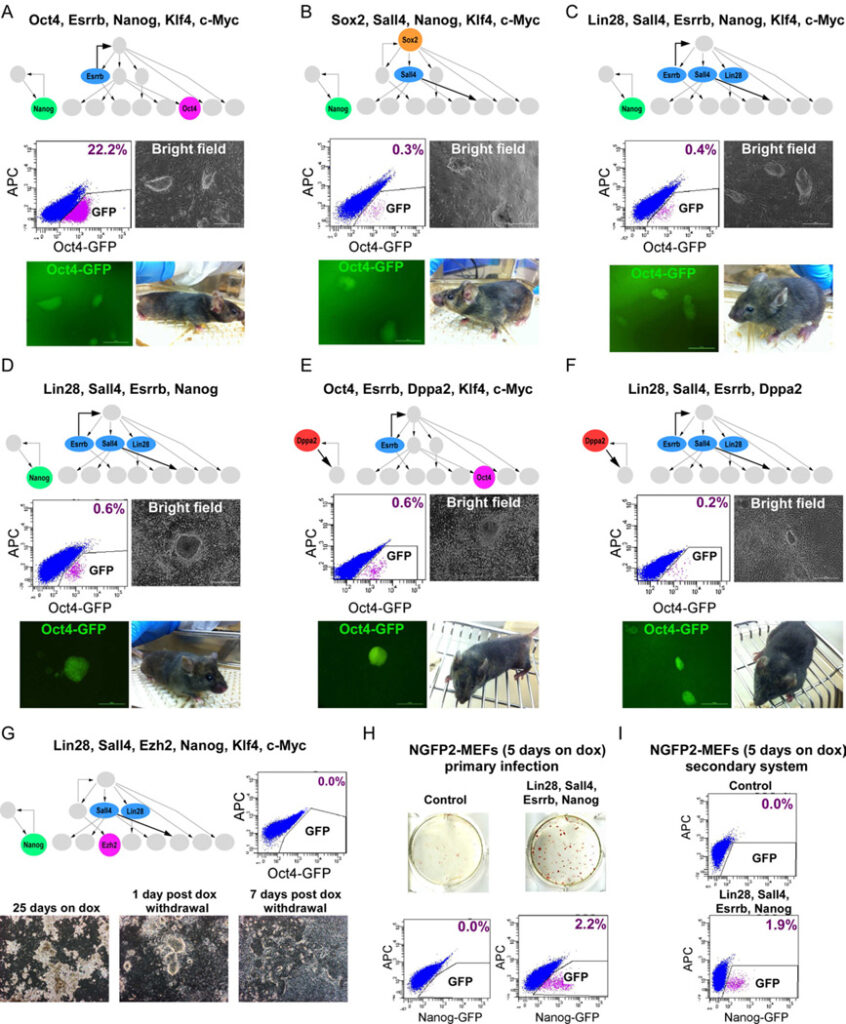
Figure 6. Cellular reprogramming with factors derived from Bayesian network
Flow cytometric analysis of GFP in Oct4-GFP cells reprogrammed with (A) Oct4, Esrrb, Nanog, Klf4, and c-Myc, (B) Sox2, Sall4, Nanog, Klf4, and c-Myc (C) Lin28, Sall4, Esrrb, Nanog, Klf4, and c-Myc (D) Lin28, Sall4, Esrrb, and Nanog, 25 days on dox, 5 days without dox. (E) Oct4, Esrrb, Dppa2, Klf4, and c-Myc (F) Lin28, Sall4, Esrrb, Dppa2, 16 days on dox, 5 days without dox. Representative images of stable dox-independent GFP+ colonies and bright-field pictures of chimeras derived from the iPSCs are shown. (G) Flow cytometric analysis of GFP in Oct4-GFP cells reprogrammed with Lin28, Sall4, Ezh2, Nanog, Klf4 and c-Myc, 7 days post dox withdrawl (upper right). Representative bright-field pictures of the cells 25 days on dox, 1 day post dox withdrawal, and 7 days post dox withdrawal are shown (bottom). (H) AP immunostaining and flow cytometric analysis of GFP in control NGFP2 MEFs (upper left) and NGFP2 MEFs reprogrammed with Lin28, Sall4, Esrrb, and Nanog by primary infection (upper right), 5 days on dox, 3 days without dox. Flow cytometric analysis of GFP is shown (bottom). (I) Flow cytometric analysis of GFP in control NGFP2 MEFs (upper) and secondary NGFP2- Lin28, Sall4, Esrrb, and Nanog MEFs (bottom), 5 days on dox, 3 days without dox. See also Figure S7.
See this image and copyright information in PMC
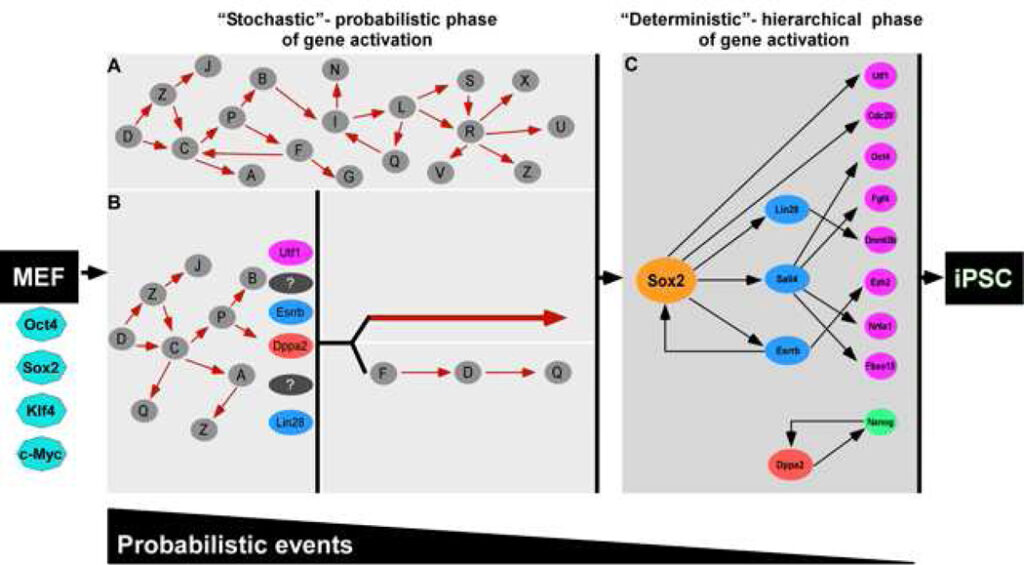
Figure 7. Two phases in reprogramming
The reprogramming process can be split into two phases: an early stochastic phase (A and B) of gene activation followed by a later more deterministic phase (C) of gene activation that begins with the activation of the Sox2 locus. After a fibroblast is induced with OSKM, the cell can proceed into either one of two stochastic phases. In A, stochastic gene activation can lead to the activation of the Sox2 locus. In B, stochastic gene activation can lead to the activation of “predictive markers” like Utf1, Esrrb, Dppa2, Lin28, which then mark cells that have a higher probability of activating the Sox2 locus. Activation of the Sox2 locus can be via two potential paths: (1) direct activation of the Sox2 locus or (2) sequential gene activation that leads to the activation of the Sox2 locus. In this model, probabilistic events decrease and hierarchal events increase as the cell progresses from fibroblast to iPSC. Solid red arrows and black arrows denote hypothetical interactions and interactions supported by our data, respectively. The white gap shown between the stochastic (A and B) and deterministic (C) panels represents the transition from induced fibroblast to iPSC illustrated between the orange dotted cluster and red cluster in Figure 2A.